Understanding chemistry of molecules through inter- and intra-molecular interactions has been the focus for scientists in the post-alchemy era. This assumes further importance as more intelligent applications of media for a desired chemical reaction demands better knowledge of solution structure, energetics and dynamics. Modern day ionic liquids (shown in Fig. 1), which are fused electrolytes near or at room temperature, possess some of the above properties and are now being extensively used for various applications.
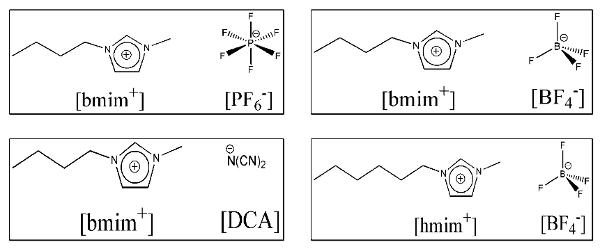
Fig. 1: Examples of some room temperature ionic liquids
Deep eutectic melts- molten multi-component mixtures at or near room temperature which is much lower than the individual melting temperature of each of the mixture components- are another set of potentially alternative media for applications in industry. Binary mixtures of organic solvents and aqueous mixtures of molecules with biological relevance are other systems of immense interest because of their relevance to both application arena and basic aspects.
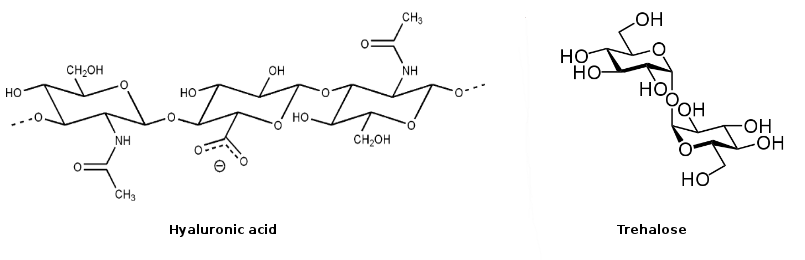
Fig. 2: Examples of biologically relevant molecules
Our principal aim is to generate considerable amount molecular level understanding of both solution structure and dynamics of these complex systems where longer-ranged interactions regulate many physicochemical and other solvent properties. For example, presence of charge-charge interaction (uijα rij-1, rij being radial distance between two charged species) leads to a very long solvation component in ionic liquids. We want to understand the reason behind this different behavior. Aqueous mixtures of polysaccharides and binary mixtures of polysaccharides and glycerol are important biologically relevant media which needs large scale simulation back up for explaining what we see via experiments.
In another activity of all atom simulation, we are focused on conformational thermodynamics of amino acids in protein (see Fig. 3). This involves calculation of thermodynamics for individual dihedrals computed from all-atom molecular dynamics simulation trajectories.
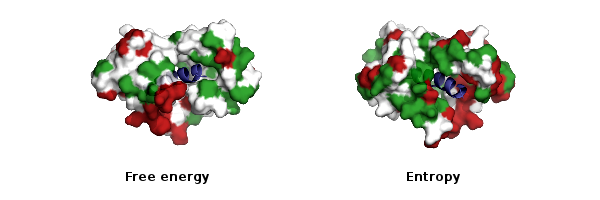
Fig. 3: Conformational thermodynamics of amino acids in protein

